Archaeogenetics
Allaby Research Group
The group has a dedicated ancient DNA laboratory. This enables us to study the evolution of crops in the Holocene directly. There are several ways in which such archaeogenetic data can be utilized. Four approaches we are taking are:
- Ancient DNA can be used to establish the presence or absence of biomarkers temporally. In this way ancient DNA can be used as a complimentary approach to phylogeography, as outlined in the example below (Ancient DNA as a temporal marker of phylogeography).
- Ancient DNA can also be used to trace directly allele evolution through time. Our current project with barley is attempting to do this.
- Palaeogenomic studies in which truly massive data sets can be generated from archaeobotanical material allowing us to directly access genome evolution in the past. Our current project with cotton is exploring this research area.
- Ancient DNA can also be used in conservation biology to identify species or genetic diversity. We actually offer an ecological forensics service to this end for bat species identification in the UK.
Bat conservation

We have recently started investigating bats in the UK through an ecological forensics service which ecologists can make use of to identify bat species on their surveys. We extract DNA from guano using ancient DNA techniques from which we can identify the species. We are also interested in learning about the genetic diversity of bat populations in the UK. If you are an ecologist interested in joining in this venture, please visit the ecological forensics page for a few more details, and/or contact Robin Allaby directly (r.g.allaby@warwick.ac.uk).
Barley

Barley was the first of the cereal crops to become domesticated, probably because its rapid growth and development suited the short spring and high summer temperatures of the early Holocene. The rapid growth of barley helps to ensure it completes its lifecycle before experiencing extreme drought stress. It is therefore an interesting crop to study for two reasons: a) it was first to be domesticated, and b) its apparent adaptation to drought stress.
We are studying barley samples from multiple strata in Qasr Ibrim, an archaeological site in Egypt, which span several thousand years. The crop here shows peculiar phenotypic traits throughout the strata that may be related to environmental conditions, such as drought stress, or genetic control. We are examining the genetic loci that determine the phenotype directly in order to help elucidate which of these two factors could have caused the change in this crop. This will help us to understand how selection under drought stress conditions in the past has helped shape this crop.
Cotton

In the archaeobotanical record, cotton species look alike. Consequently, ancient DNA techniques can inform us a great deal about the evolution of cotton domestication. For instance, the cotton present at Qasr Ibrim may be Gossypium herbaceum, or G. arboreum. These crops may look similar, but one originates on the African continent and the other on the Indian subcontinent, consequently a knowledge of which was used would inform us a great deal about early cotton usage and origins.
The Gossypium genus has been undergoing genome expansion through retrotransposon activity in most of its genomic lineages, and there has been some suggestion that the process is ongoing. We are applying the new technique of palaeogenomics to look for signs of genome change over the past few thousand years, including retrotransposon activity and evidence for selection.
Ancient DNA as a temporal marker of phylogeography
Brown, Allaby et al (1998) discovered evidence for a hitherto unseen G genome expansion of wheats out of the Near East by finding G genome specific alleles of the high molecular weight glutenin gene in 3300 year old wheat remains from Assiros in Greece:
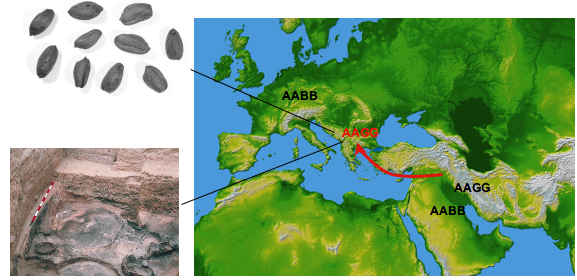
Until then it was thought that the only tetraploid wheat to move from the Near East to Europe was of the AABB genome type, such as emmer, the discovery of an AAGG type tetraploid in Europe seemed incredible at the time. However, archaeobotanical evidence soon supported the finding (Jones et al 2000 Veg. Hist. Bot. 133-146), supporting the discovery of an extinct expansion of a wheat lineage into Europe.
Ancient DNA publications
Palmer SA, Moore JD, Clapham AJ, Rose P, Allaby RG (2009) Archaeogenetic Evidence of Ancient Nubian Barley Evolution from Six to Two-Row Indicates Local Adaptation. Plos One 4(7): e6301. doi:10.1371/journal.pone.0006301
Ho, S.S.W., Kolokotronis, S.-O., Allaby R.G. (2007) Elevated substitution rates estimated from ancient DNA sequences. Biology Letters doi:10.1098/rsbl.2007.0377.
Freitas F.O., Bendel G., Allaby R.G., and Terence A. Brown (2003) DNA from Primitive Maize Landraces and Archaeological Remains: Implications for the Domestication of Maize and its Expansion into South America. Journal of Archaeological Science 30:901-908.
Allaby R.G., Banerjee M., Brown T.A. (1999). Evolution of the high molecular weight glutenin loci of the A,B,D and G genomes of wheat. Genome 42:296-307.
Brown T.A., Allaby R.G., Sallares R., Jones G. (1998). Ancient DNA in charred wheats: taxonomic identification of mixed and single grains. Ancient Biomolecules 2:185-193.
Allaby R.G., O'Donoghue K., Sallares R., Jones M.K., Brown T.A. (1997) Evidence for the survival of ancient DNA in charred wheat seeds from European archaeological sites. Ancient Biomolecules
Brown K.A., Allaby R.G., Garland A.N., Brown T.A. (1996). Ancient DNA in archaeological bones and plant remains. In Proceedings of the Archaeological Sciences Conference 1991.
Allaby R.G., Jones M.K., Brown T.A. (1994). DNA in charred wheat grains from the Iron Age hill fort at Danebury, England. Antiquity 68:126-32.
Brown T.A., Allaby R.G., Brown K.A., O'Donoghue K., Sallares R. (1994). DNA in wheat seeds from European archaeological sites. In Conservation of Plant Genes II: Intellectual property rights and DNA utilisation (eds. Adams R.P. et al) 37-46. Missouri Botanical Gardens, St.Louis.
Brown T.A., Allaby R.G., Brown K.A., O'Donoghue K., Sallares R. (1994). DNA in wheat seeds from European archaeological sites. Experientia 50:571-575.
Brown T.A., Allaby R.G., Brown K.A., Jones M.K. (1993). Biomolecular archaeology of wheat: past, present and future. World Archaeology 25:64-7
