Zak Ogi-Gittins
About me & Research Interests
Hello! I am a 1st year PhD student at the Mathematics for Real-World Systems Centre for Doctoral Training. Since joining the MathSys CDT, I have become interested in Mathematical Epidemiology, especially given the attention that the field has gained during the COVID-19 pandemic. I am also interested in applied dynamical systems and parameter inference.
Academic Projects
MathSys Group Project: "Adaptive management during an ongoing pandemic"
I worked within a small group of MathSys students to evaluate the impact of using Adaptive Management techniques to improve decision making during an epidemic. The logic underpinning Adaptive Management is that parameter uncertainty is likely to be resolved as an outbreak unfolds. Mathematically, we can use the information that our parameter knowledge will change in the future to aid our decision-making processes today. Ideally, these strategies would be used in present-day epidemic modelling but significant work is required for this approach to be more popular in the modelling community.
In our project, we attempted to evaluate the optimum time that the UK should have begun lockdown (given the up to date knowledge at the time) using Adaptive Management methodology. We found (as is well documented in the literature) that the decision-making process is highly dependent on the loss function, i.e. a linear combination of lives lost and time spent in lockdown.
MathSys Masters Dissertation: "How mis-specifying time-varying generation intervals affects reproduction number inference"
Statistical monitoring of epidemics relies on the data streams that are provided. These streams suffer multiple biases, i.e. delay-times, under-reporting, etc. In spite of these biases, there are several methods that can eliminate some of the issues when inferring the time-dependent reproduction number during an ongoing epidemic [Cori et al, Thompson et al., Epi-Estim]. In this report, we investigated the effects of generation-interval mis-specification on reproduction number inference, and which properties of misspecification still enable the user to process whether the inference is likely to be an over/under-estimate.
Highly Pathogenic Avian Influenza Rapid Response
During the UK’s H5N1 (bird-flu) outbreak, a small group within SBIDER (Systems Biology & Infectious Disease Epidemiology Research) began modelling the spread between poultry holdings across England, Wales and Scotland. This piece of work uses a spatial model established by Keeling et al., which enables a significant improvement in the speed of computation.
From January-March 2022, we conducted an initial analysis using MCMC algorithms to estimate the dispersion kernel present in the model, as well as other key parameters such as transmissibility and susceptibility parameters. From this, we were able to simulate counterfactual scenarios to evaluate the relative risk of the outbreak that occurred in North Yorkshire. We also modelled the impact of ring-culling and enhanced surveillance.
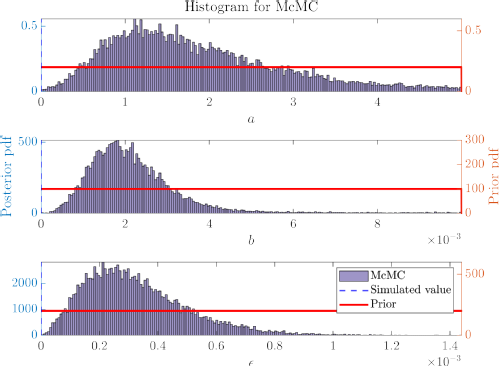
Since then, I have worked on extending this initial project to look at different models, including a spark-term model, which accounts for wild bird incursions (see posteriors for fitted parameters below). We plan to compare the models that we have fitted and continue our counterfactuals analysis.
Reproduction Number Inference using Weekly Incidence Data
We are currently working with an external collaborator on a project that enables basic incidence projection, as well as improving reproduction number inference. One of the real-world problems with reproduction number inference is that incidence reports are often collated per week, by health organisations, which suggests we should also make use of serial intervals (distribution of delays between infector and infected report times) resolved on a per-week basis. This is problematic for inference methods such as those used in EpiEstim [https://shiny.dide.imperial.ac.uk/epiestim/Link opens in a new window] because these methods assume that no infector-infectee pairs coexist in the same calendar week. The ideal solution is to use the natural daily serial intervals and to produce reproduction number inference by disaggregating weekly incidence reports. We plan to develop and test at least one novel method that implements incidence disaggregation and subsequently compare this reproduction number inference with existing methods.
Teaching Responsibilities
In my first year, I was a TA on the MA398 and MA4M1 modules in the Warwick Maths Department
Contact Information
Office: D2.11 Complexity Science
Zeeman Building,
University of Warwick,
Coventry,
CV4 7AL