Abdnacer ("Nacer") Bouchekhima
PhD Title: Quantification of The Plant Endoplasmic Reticulum
Supervisors: Lorenzo Frigerio & Markus Kirkilionis
The endoplasmic reticulum of higher-plant cells consists of a relatively immobile polygonal network of tubules and cisternae located close to the plasma membrane (cortical ER), together with a system of highly mobile ER elements distributed throughout the cytoplasm. The two ER systems are interconnected. Close to the cortical ER is the Golgi which is very dynamic in its movement. Both compartments are part of the plant’s secretory pathway. Communication between compartments is by vesicular traffic. One of the major aims in this field of research is to understand the routes and timescales with which proteins are transcribed, transported and sorted in the secretory pathway and finally stored in the vacuoles. This is of immense importance for biotechnology where transgenic plants are used to produce pharmacologically active compounds.
The ER of higher-plant cells consists of a relatively immobile polygonal network of tubules and cisternae located close to the plasma membrane (cortical ER), together with a system of highly mobile ER elements distributed throughout the cytoplasm. The two ER systems are interconnected. Close to the cortical ER is the Golgi (labelled red) which is very dynamic in its movement. Both compartments are part of the plant’s secretory pathway. Communication between compartments is by vesicular traffic. One of the major aims in this field of research is to understand the routes and timescales with which proteins are transcribed, transported and sorted in the secretory pathway and finally stored in the vacuoles. This is for example of immense importance for biotechnology where transgenic plants are used to produce pharmacologically active compounds.
We concentrate on three main parts: Static ER, Dynamic ER, and FRAP simulation in the ER.
Static ER:
One of the most difficult obstacles to make biological sciences more quantitative is the lack of understanding the interplay of form and function. Each cell is full of complex shaped objects which moreover change their form over time. To tackle this problem we suggest the use of geometric invariants which are able to produce precise reference points to compare the cell's different functional elements like organelles under fixed and varying physiological conditions. This can be achieved by exploiting the power of Confocal imaging which opened new horizons to the scientific community by enabling live imaging of different sub-cellular processes. This study enables the broadening of our knowledge on how different physiological characteristics of organelles are influenced by their geometry.
Details of this study can be found in ...
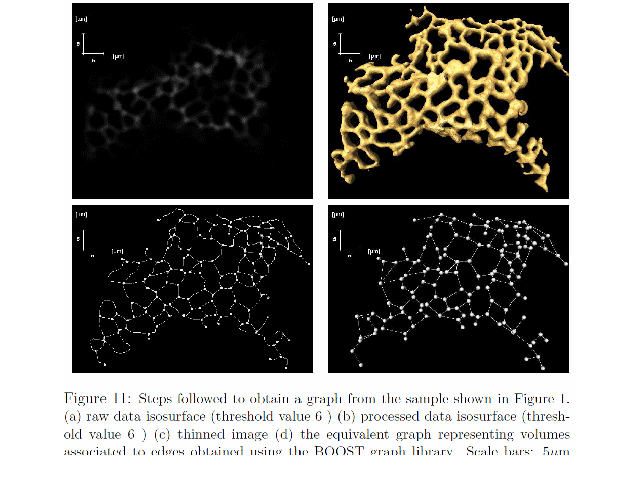 |
Dynamic ER:
-
Pre-filtering using the Median filter: To balance between noise minimazation and preservation of as much information as possible in the original images; we choose to uniquely use the Median filter with the smallest possible kernel (3x3). This is because of the heavy presence of Salt-and-Pepper noise (refer to the movies) in the data. This is in contrast with the Median -Gaussian smoothing chain proposed earlier in our GQER paper.
- Enhancement using CLAHE: A common feature in confocal micrographs is that only the portions of the sample that are in the focus plane will show the signal (GFP in our case); so in order to obtain a clear 2-D image of the ER it is usually necessary to scroll through the sample to find a portion that is aligned in the plane of focus. Even so the signal strength varies from one part of the image to another because the ER does not completely lie in the same z-plane (due the pressure exercised by the vacuole on the ER,) or because GFP is not uniformly distributed throughout the ER). To even out the distribution of used grey values, make hidden features of the ER more visible, and more importantly to simplify the background removal operation, we follow the above Median filter operation by a Contrast Limited Adaptive Histogram Equalisation (CLAHE) enhancement step. CLAHE partitions animage into smaller contextual regions and applies the histogram equalisation to each one in turn. So rather than working on the entire image, CLAHE operates on small contextual regions in the image. Each regions's contrast is enhanced, so that the histogram of the output region approximately matches the histogram specified by a certain 'Distribution' (we used Rayleight because it yields 2 bell shaped histogram populations). The method then combines the neighbouring regions using bilinear interpolation in order to eliminate artificially induced boundaries.
- Background removal Using Sauvola LocalAdaptive algorithm: Now that we have a clearer the image with maximum information conserved, we apply state of the art Sauvola local Adaptive thresholding technique, which treats the image as a set of connected regions (or windows of size w x w centred around pixel(x,y) ), and associate a different threwhold to each one in turn, taking into account the local mean and the local variance of the pixel intensities, in contrast to rival methods such as the mean local adaptive thresholding method, that take only the mean. This is an other vital step to highlight details (holes) and avoid false edges (very close edges). The smaller the window size the more details we get.
- To increase the speed of computations and make them more efficient we use Integral and Square imagesthat can be obtained by a single pass. We implemented this part in matlab and found it it extremely fast!
- Thresholding using the Two Peaks approach: applying CLAHE and removing the background yields two distinct cluster populations, which have Gaussian histograms; that is a foreground and a background clusters of pixels. A naive, yet efficient, way to separate these populations is to take the average of the two peaks of these Gaussian distributions as a threshold value. A more elaborate approach is to use the Minimum Error Global Thresholding method where the assumption that the image histogram contains two Gaussian distributions is exploited to turn the thresholding issue into minimazation problem. A major draw back to this approach is that it depends on the initial value. We found that by varying the initial value we get conflicting results which led us to simply use the two peaks mean approach.
- SkeletonizationSkeletons are vital for associating physical and geometrical parameters to the network. There are several skeletonization algorithms to choose from and the least time consuming and most convenient we found is: The Euclidean Sequential voxel based Thinning algorithm (More efficient methods, such as skeleton extraction by mesh contraction, were avoided because (1) this is not the main aim of this investigation and (2) most importantly they produce the same results) to extract the skeleton of the network.
- The skeleton is a simplification of the real network and plays a critical role in the calculations to come.
- To avoid unnecessary complexities in motion analysis we decide to work in the skeleton domain (This is an image that only contains the skeleton and nothing else!). We Further simplify the problem by isolating the skeleton junctions (Where edges meet) and remove all other points (Inner points)
- Finally we highlight the remaining points by circles of known radii to further facilitate tracking and motion analysis.
- Step 9 is the beginning of the next phase of this study, that is Motion analysis.....
- .... to be continued ...More to follow on how to evaluate the network motion...
Simulation of Protein Diffusion in the ER (1D & 2D)
Details to follow shortly
Mathematical modeling:
The ER will be geometrically characterised and subsequently discretised as a transport network. This will first be done by a 1D representation of the network branches. On each branch a diffusion equation will be solved and characteristic distribution times of proteins be calculated. Diffusion coefficients will directly be measured for differently sized proteins with the help of confocal imaging. Here the effects of the network topology on effective transport times will be a major topic in research. Next we will characterise the ER cisternae and tubes by more sophisticated 2D and 3D discretisations, in close cooperation with the imaging working groups at the computer science department. This will alow to investigate hypothesis about molecular crowding in the ER lumen. If successful we will at the end of the project study transport from the ER to other compartments. This will involve the mathematical modelling of vesicular traffic.
In this part we characterise the transport rates and routes of the secretory pathway in an interplay between confocal imaging and mathematical modelling with transport equations.
Methods used for confocal imaging:
We will use phtoactivatable green fluorescent proteins (PA-GFP) to study transport of proteins, first in the ER. This will be done both in pulse experients for proteins bound to the ER membrane, but also in the ER lumen. The technique is to track the movement of the activated PA-GFP fusions. Thus photoactivation will provide invaluable information on a number of parameters:
- the route(s) followed by the protein in the ER network
- the timing of the transport event;
- the half-life of the protein.
Such parameters have so far proven very difficult to determine using standard biochemical methods such as pulse-chase analysis and cell fractionation, or ‘static’ methods such as immunocytochemistry. Likewise, existing live imaging methods such as FRAP, which use standard fluorescent proteins, are not suitable. The unique advantage of using plant epidermal cells for this study is that the presence of a large central vacuole results in all organelles being confined to a thin cortical layer beneath the plasma membrane. This can be studied as a bidimensional plane by confocal microscopy, thus simplifying the pixel-by-pixel analysis of protein dynamics with time.
