Materials and Methods
FtsZ
FtsZ had been expressed and purified during mini-project 1 using an established protocol (Mukherjee and Lutkenhaus 1998), except that the cells were lysed by sonication (Beuria, Krishnakumar et al. 2003) rather than by using a French press. Two batches were processed, the first from 1 L of cell culture producing 8 mL FtsZ at a concentration of 3.8 mg/mL (Batch A), and the second from 4 L cell culture producing 10 mL FtsZ at a concentration of 7.77 mg/mL (Batch B). FtsZ was stored in a buffer (Buffer A) consisting of 50 mM KCl, 1mM EDTA, 50mM Tris buffer: pH 7.9 and 10% glycerol in 1mL and 100 μL aliquots at –80 °C.
Some experiments were performed with the FtsZ kindly donated by Raul Pacheco-Gomez (Batch RPG) for comparison purposes.
The standard solution for investigating FtsZ polymerisation using right-angled light scattering or linear dichroism (the polymerisation buffer) consisted of 50 mM MES buffer, pH 6.5 unless otherwise specified, 50 mM KCl, 10 mM Mg Cl2, and 11 μM FtsZ. Salt and MES solutions had been filtered to 0.2 μM. A batch of solution, without FtsZ, was made up each day such that the concentrations would be correct once the FtsZ was added, and refiltered to 0.2 μm by expressing the solution through a syringe mounted filter. . The second filtering was found to reduce the variation with time of the light scattering measurement. In later experiments the buffer was degassed by sucking the buffer into a 10 mL Luer fitting syringe, sealing the end while excluding as much air as possible, and then drawing the plunger and tapping on the side of the syringe to induce dissolved gases to come out of solution into the partial vacuum that is formed
Right angled light scattering
Right angled light scattering is an established technique for monitoring the assembly and disassembly of FtsZ protofilaments (Mukherjee and Lutkenhaus 1999). Right angled light scattering was measured using a Perkin Elmer LS50b Fluorescence Spectrometer, using a triple quartz window black cuvette (Stama Ltd) with a nominal volume of 50 μL and a path length of 3 mm. Measurements were made at 450 nm and the excitation and emission slit widths were set at 2.5 nm. The cuvette was washed three times with water and once with ethanol and air dried prior to use. Sufficient FtsZ solution and polymerisation buffer to make up 100 μL solution with 11 μM FtsZ concentration was mixed in an eppendorf by gently sucking and expelling once from a pipette, and then gently transferred by pipette to the cuvette.
The right angled light scattering was monitored until the value had stabilised and then the cuvette was removed, 1 μL of 20 mM GTP was pipetted into the cuvette added to initiate polymerisation. It was mixed by gently sucking the mixture once into a 100 μL pipette and expelling it back into the cuvette.
Polymerisation was performed at room temperature, although other protocols (e.g. Beuria, Krishnakumar et al. 2003) specify 37 °C.
Analytical Ultracentrifugation (AUC) analysis of proteins.
AUC is a well established technique for determining the relative concentrations of components in solution. The density variation of a solution is monitored as macromolecules drift outwards at different rates when a solution is centrifuged. The time varying density is then analysed to derive data on the distribution of different sizes of marcromolecules present. By making assumptions about the physical characteristics of these molecules it is possible to interpret the data in terms of the distribution of molecular weights (Edelstein and Schachman 1967). A sample of Batch A was subjected to analysis by AUC using a Beckham Coulter XL-A analytical ultracentrifuge kindly made available by Birmingham University.
Linear dichroism
Linear dichroism (Dafforn, Rajendra et al. 2004) has been introduced as an additional technique for monitoring the structure and kinetics of FtsZ protofilament formation (Marrington, Small et al. 2004)
Linear dichroism was measured using the LD cell with either a central stationary quartz rod or the new nested capillary cell described in section 2, mounted on a Jasco J-715 spectropolarimeter adapted for LD spectroscopy. Spectral and kinetics data was gathered using Jasco software. Kinetics data was smoothed if required in order to see the underlying change in the mean using a two identical recursive digital filters, where the output is related to the input
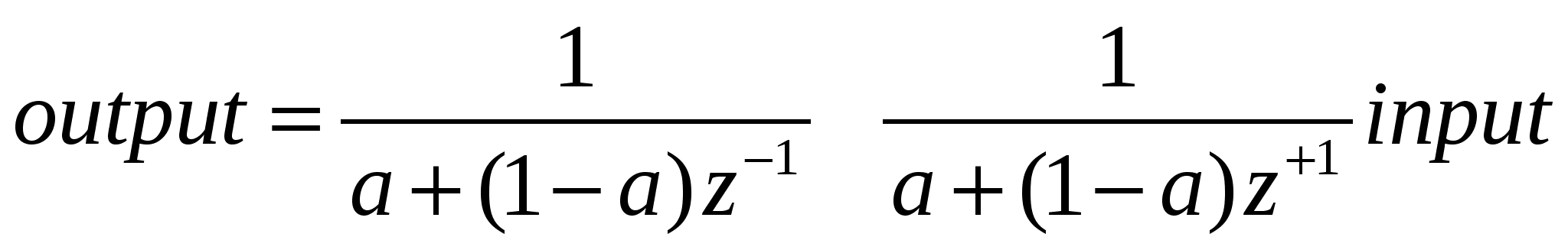
This was implemented using Microsoft Excel®, and has the advantage that it is simple to implement and has no group delay, where the group delay is an indication of the shift in time of the data as a result of filtering. This filtering was only applied when the signal was relatively constant and not when the signal was changing quickly with time, to avoid hiding valuable transient data.
Unless otherwise stated measurements were made with the voltage of the motor which drives the outer capillary at 3V when used with the central quartz rod, and 2.5V when used with the new nested capillary. The voltage was reduced with the new system because the inner capillary seemed to rotate more smoothly as indicated by the sound it made whilst rotating, and it was assumed that this would correspond to a more regular Couette flow.
Sonication
Sonication was used as a standard way of subjecting the solution to mechanical stress in a relatively repeatable way. It was done by placing the solution into a 0.5 mL Eppendorf Micro Test Tube and then placing the tube in an expanded polystyrene holder floating on the surface of an uncalibrated sonicating bath filled with water at room temperature.
Molecular structure analysis
Surface electrostatic potentials were calculated and visualised using the DeepView /Swiss-PdbViewer version 3.7. http://www.expasy.org/spdbv/ (Guex and Peitsch 1997; Schwede, Kopp et al. 2003) and were either rendered using POV-Ray for Windows Version 3.6.1 or by rendering in Solid3D using open GL and taking a screen snapshot.
Residue conservation between protein homologues was calculated using the ConSurf Server 3.0 http://consurf.tau.ac.il/index.html (Glaser, Pupko et al. 2003)and visualised using First Glance in Jmol (http://firstglance.jmol.org)
